Scheldt species taxon details
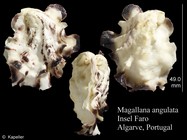
Magallana angulata (Lamarck, 1819)
1039387 (urn:lsid:marinespecies.org:taxname:1039387)
accepted
Species
Crassostrea angulata (Lamarck, 1819) · unaccepted
marine
(of ) Lamarck, [J.-B. M.] de. (1819). <i>Histoire naturelle des animaux sans vertèbres</i>. Tome sixième, 1re partie, vi + 343 pp. Paris, published by the author. , available online at http://www.biodiversitylibrary.org/item/47441
page(s): 198 [details]
page(s): 198 [details]
MolluscaBase eds. (2025). MolluscaBase. Magallana angulata (Lamarck, 1819). Accessed through: VLIZ Consortium Scheldt Species Register at: https://scheldemonitor.nl/speciesregister/aphia.php?p=taxdetails&id=1039387 on 2025-09-12
VLIZ Consortium. Scheldt Species Register. Magallana angulata (Lamarck, 1819). Accessed at: https://www.scheldemonitor.be/speciesregister/aphia.php?p=taxdetails&id=1039387 on 2025-09-12
Date
action
by
original description
(of ) Lamarck, [J.-B. M.] de. (1819). <i>Histoire naturelle des animaux sans vertèbres</i>. Tome sixième, 1re partie, vi + 343 pp. Paris, published by the author. , available online at http://www.biodiversitylibrary.org/item/47441
page(s): 198 [details]
basis of record Salvi D. & Mariottini P. (2017 [nomenclatural availability: 2016]). Molecular taxonomy in 2D: a novel ITS2 rRNA sequence-structure approach guides the description of the oysters' subfamily Saccostreinae and the genus <i>Magallana</i> (Bivalvia: Ostreidae). <em>Zoological Journal of the Linnean Society.</em> 179(2): 263-276., available online at https://doi.org/10.1111/zoj.12455
page(s): 5, 9; note: new combitantion herein; Salvi & Mariottini (2016) treat Crassostrea angulata as a synonym of C. gigas but include all Asian Pacific species in Magallana [details]
page(s): 198 [details]
basis of record Salvi D. & Mariottini P. (2017 [nomenclatural availability: 2016]). Molecular taxonomy in 2D: a novel ITS2 rRNA sequence-structure approach guides the description of the oysters' subfamily Saccostreinae and the genus <i>Magallana</i> (Bivalvia: Ostreidae). <em>Zoological Journal of the Linnean Society.</em> 179(2): 263-276., available online at https://doi.org/10.1111/zoj.12455
page(s): 5, 9; note: new combitantion herein; Salvi & Mariottini (2016) treat Crassostrea angulata as a synonym of C. gigas but include all Asian Pacific species in Magallana [details]
 Present
Present  Inaccurate
Inaccurate  Introduced: alien
Introduced: alien  Containing type locality
Containing type locality
| Language | Name | |
|---|---|---|
| Dutch | Portugese oester [from synonym] | [details] |
| English | Portuguese oyster [from synonym] | [details] |
To Barcode of Life (3 barcodes)
To Barcode of Life (338 barcodes) (from synonym Crassostrea angulata (Lamarck, 1819))
To Biodiversity Heritage Library (32 publications) (from synonym Crassostrea angulata (Lamarck, 1819))
To European Nucleotide Archive, ENA (Crassostrea angulata) (from synonym Crassostrea angulata (Lamarck, 1819))
To European Nucleotide Archive, ENA (Magallana angulata)
To GenBank (137543 nucleotides; 50519 proteins) (from synonym Crassostrea angulata (Lamarck, 1819))
To GenBank (3 nucleotides; 3 proteins)
To Malacopics (Magallana angulata (Lamarck,1819) France, Bretagne, Finistère, Terenez)
To PESI (from synonym Crassostrea angulata (Lamarck, 1819))
To Barcode of Life (338 barcodes) (from synonym Crassostrea angulata (Lamarck, 1819))
To Biodiversity Heritage Library (32 publications) (from synonym Crassostrea angulata (Lamarck, 1819))
To European Nucleotide Archive, ENA (Crassostrea angulata) (from synonym Crassostrea angulata (Lamarck, 1819))
To European Nucleotide Archive, ENA (Magallana angulata)
To GenBank (137543 nucleotides; 50519 proteins) (from synonym Crassostrea angulata (Lamarck, 1819))
To GenBank (3 nucleotides; 3 proteins)
To Malacopics (Magallana angulata (Lamarck,1819) France, Bretagne, Finistère, Terenez)
To PESI (from synonym Crassostrea angulata (Lamarck, 1819))